Here, you will find a short selection of analysis methods, which I can offer you. Just choose your method an click to see a short description and example pipelines.
I want to point out, that those analysis pipelines are only examples and can be adapted to your needs. This means, that I can change programs within the pipeline, adapt or add steps according to your wishes and even I can change the version numbers of the programs within the pipelines. If you want to find out why this might be necessary to secure analysis reproducibility please click here.
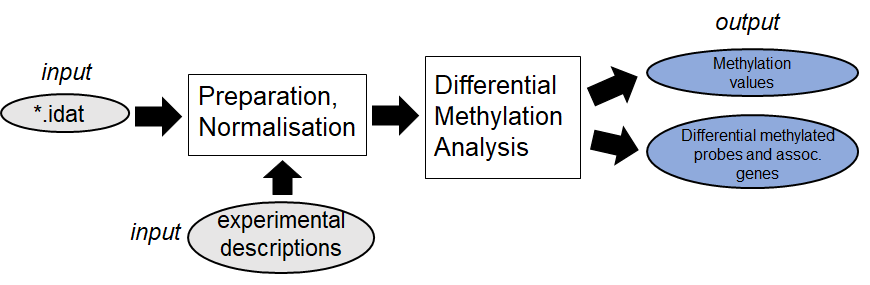
Methylation Analysis (EPIC):
Raw-data idat files will be processed with the R package minfi.
After normalisation, the methylation value per locus and the differential methylated probes
between two experimental conditions will be calculated. (see
pipeline
)
close window
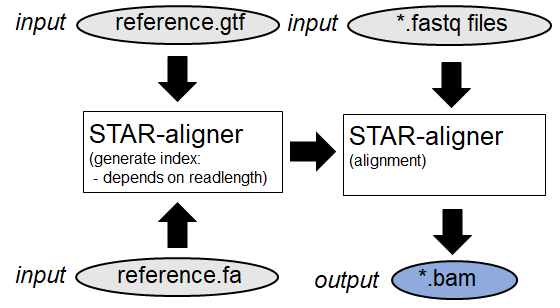
RNA sequencing analysis:
Raw-data fastq files will be aligned against a given reference. (Example:
STAR
)
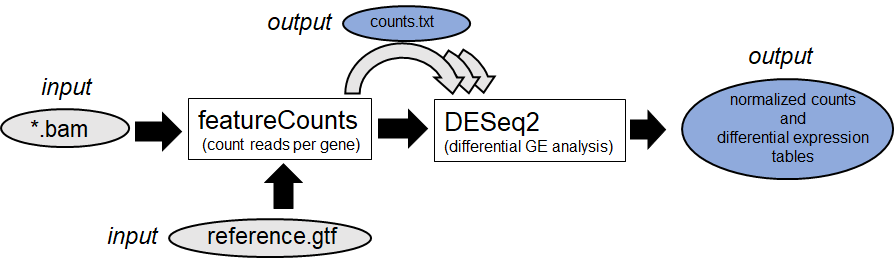
Afterwards several downstream methods are possible.
1. Differential Gene Expression Analysis (Example:
DESeq2
)
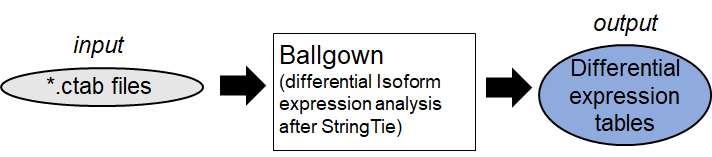
2. Differential Isoform expression Analysis (Example:
Ballgown
)
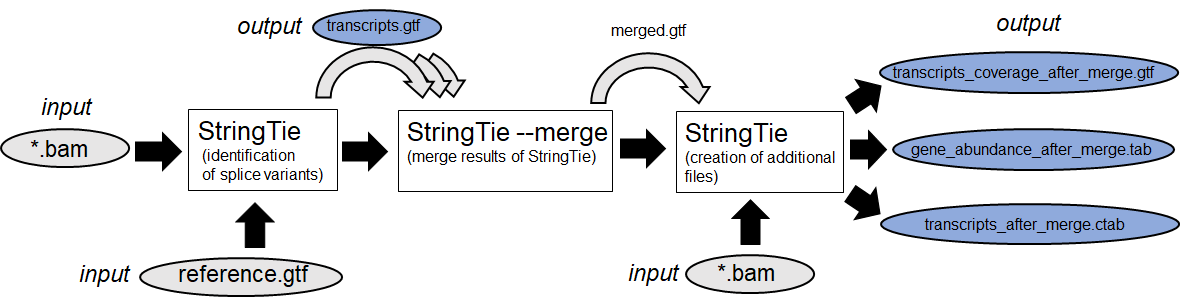
3. Identification of new Isoforms (Example:
Stringtie
)
close window
ATAC sequencing analysis:
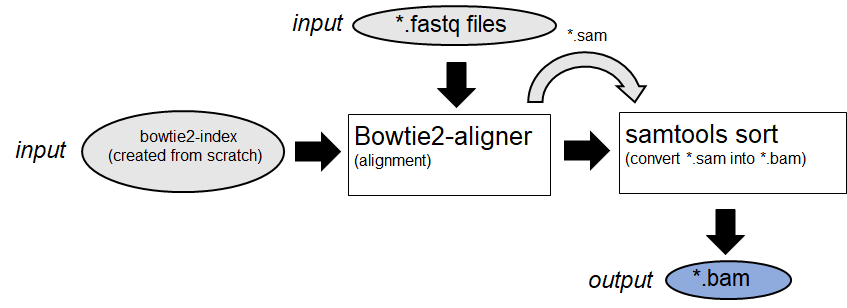
Raw-data fastq files will be aligned against a given reference. (Example:
Bowtie2
)
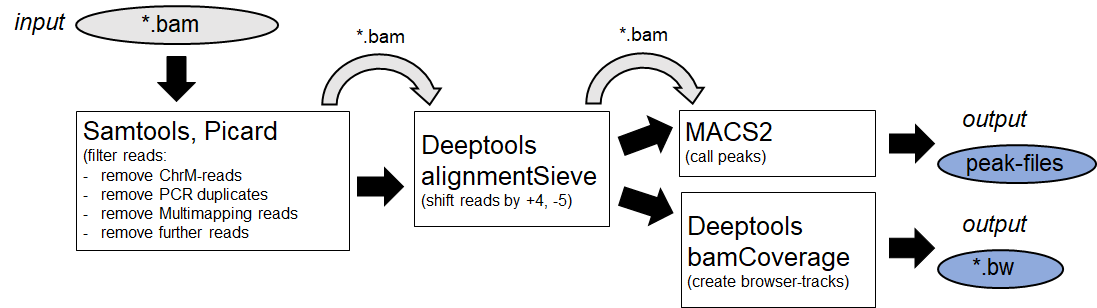
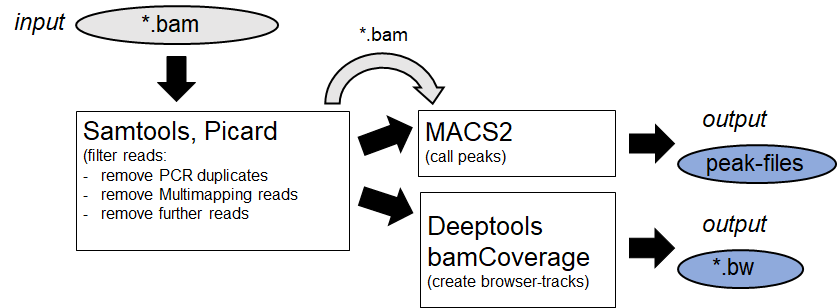
Afterwards the alignemts are filtered by specific features and Peaks are identified. (Example:
MACS2
)
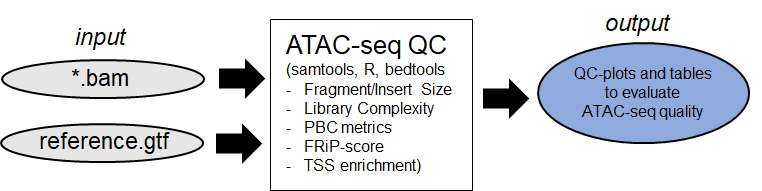
Additionally the ATAC-seq results can be checked for specific quality metrices (see
ATAC-seq quality control
)
Finally, Further downstream approaches are possible.
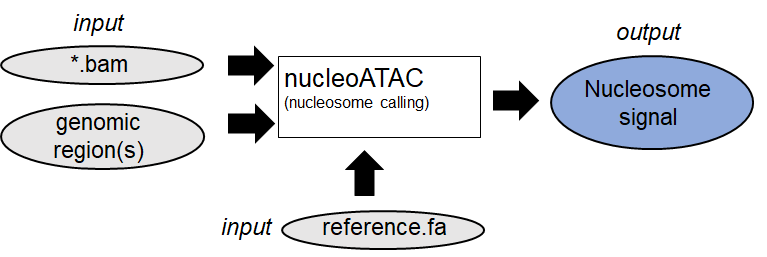
1. Identification of Nucleosome positions (Example:
nucleoATAC
)
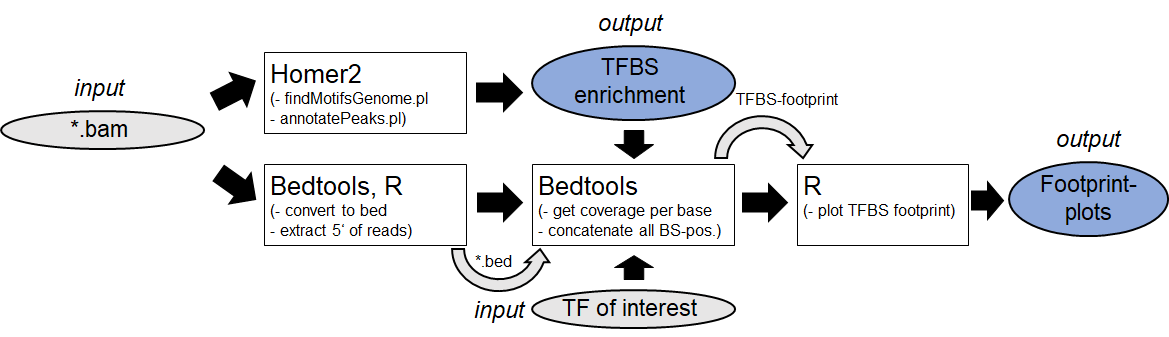
2. TFBS-enrichment analysis and footprinting (Example:
Homer
)
3. Differential Peak analysis
close window
ChIP sequencing analysis:
Raw-data fastq files will be aligned against a given reference. (Example:
Bowtie2
)
Afterwards Peaks are identified. (Example:
MACS2
)
Finally, Further downstream approaches are possible.
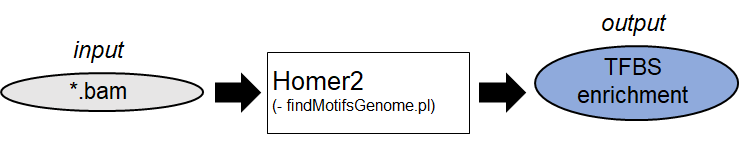
1. TFBS-enrichment analysis and Motif Discovery (Example:
Homer
)
2. Differential Peak analysis
close window
Other methods:
If you miss a certain method, or if you want to adapt a method to your special wishes, just
contact me and we can discuss together what I can do for you.
close window
return to description
close window
return to description
close window
return to description
close window
return to description
close window
return to description
close window
return to description
close window
return to description
close window
return to description
close window
return to description
close window
return to description
close window

return to description
close window
return to description
close window
return to description
close window
0













