Bioinformatics.Expert UG (haftungsbeschränkt)
Fast, precise, reproducible, flexible and cost effective data analysis.

Hi! My name is Felix and I want to support you with my expertise in bioinformatic data analysis.
Go to “Services” to see my scope and how I can help you today.
Visit “Pipelines” if you want to see a short selection of methods, which I can help you to analyze.
Those include e.g. the analysis of DNA methylation arrays but also NGS-based approaches.
Find out, how different analysis methods influence bioinformatic results and how I can help you to ensure reproducibility to your methods and outcome. (click on the image)
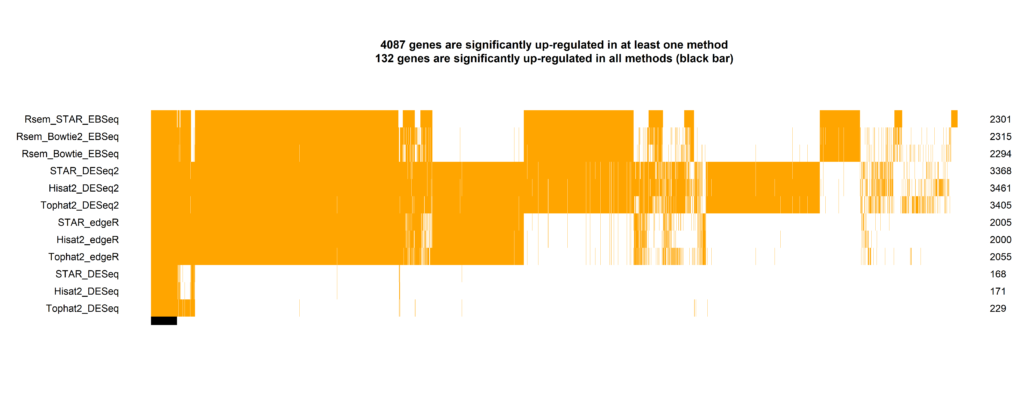
Contact me if you have questions or if you are interested in this service and also if you want to give feedback.
Scientific contributions:
Bioinformatics.Expert UG as Co-Author:
Waldbillig F, Bormann F et al. An m6A-Driven Prognostic Marker Panel for Renal Cell Carcinoma Based on the First Transcriptome-Wide m6A-seq, Diagnostics 2023 Feb 21;13(5):823 2023
>>Data analysis and visualisation
Simon T, Riemer P et al., DNA methylation reveals distinct cells of origin for pancreatic neurendocrine carcinomas and pancreatic neuroendocrine tumors, Genome Medicine 2022 (14:24)
>> DNA-methylation analysis (EPIC-chips)
Jäger P, Geyh S et al., Acute myeloid leukemia-induced functional inhibition of healthy CD34+ hematopoietic stem and progenitor cells, Stem Cells. 2021 Sep 39(9);1270-1284.
>> RNAseq-analysis and visualisation
Chteinberg E, Vogt J, Kolarova J, Bormann F et al., The curious case of Merkel cell carcinoma: epigenetic youth and lack of pluripotency., Epigenetics. 2020 May 30;1-6.
>> RNAseq-analysis and visualisation
Bioinformatics.Expert acknowledged:
Bashir S, Dang T et al., Enhancement of CRISPR-Cas9 induced precise gene editing by targeting histone H2A-K15 ubiquitination, BMC Biotechnol. 2020 Oct 23;20(1):57. doi: 10.1186/s12896-020-00650-x
>> Development of a pipeline for Amplicon-sequencing analysis (Please get in contact, if you want to use it)
